Abstract
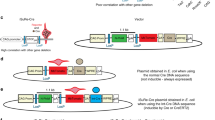
Chromosomal deletions are a common feature of epithelial tumours and when further defined by homozygous deletions, are often the location of tumour suppressor genes. Deletions within the short arm of chromosome 3 occur very frequently in human carcinomas: a minimal region of loss at 3p21.3 (the Luca) region has been defined by overlapping homozygous deletions in lung and breast cancer cell lines. Using a rapid strategy for Cre-loxP chromosome engineering, a deletion of approximately 370 kb was created in the mouse germline corresponding to the deleted region at 3p21.3. The deletion when homozygous is embryonic lethal. Heterozygotes develop normally despite being haplo-insufficient for twelve genes including the candidate tumour suppressor gene Rassf1. Because damage to 3p21.3 often occurs very early in the sequence of genetic changes that lead to malignancy, particularly in lung and breast cancer, further genetic damage to these mice will provide the opportunity to model multi-step tumorigenesis of these tumours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, Hosoe S, Lerman MI, Minna JD, Maher ER, Latif F . 2001 Oncogene 20: 1509–1518
Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD . 2001 J. Natl. Cancer Inst. 93: 691–699
Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG . 2001 Cancer Res. 61: 7034–7038
Chung GT, Sundaresan V, Hasleton P, Rudd R, Taylor R, Rabbitts PH . 1995 Oncogene 11: 2591–2598
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer G . 2000 Nat. Genet. 25: 315–319
Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER . 2001 Proc. Natl. Acad. Sci. USA 98: 7504–7509
Hesketh R . 1997 The oncogene and tumour suppressor gene fact book Academic Press
Justice MJ, Zheng B, Woychik PR, Bradley A . 1997 Methods 13: 423–436
Kondo M, Ji L, Kamibayashi C, Tomizawa Y, Randle D, Sekido Y, Yokota J, Kashuba V, Zabarovsky E, Kuzmin I, Lerman M, Roth J, Minna JD . 2001 Oncogene 20: 6258–6262
Kok K, Naylor SL, Buys CHCM . 1997 Adv. Cancer Res. 72: 27–92
Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG . 2001 Cancer Res. 61: 6688–6692
LePage DF, Church DM, Millie E, Hassold TJ, Conlon RA . 2000 Proc. Natl. Acad. Sci. USA 97: 10471–10476
Lerman MI, Minna JD . 2000 Cancer Res. 60: 6116–6133
Li Z-W, Stark G, Gotz J, Rulicke T, Muller U, Weissmann C . 1996 Proc. Natl. Acad. Sci. USA 93: 6158–6162
Macleod K . 2000 Curr. Opin. Genet. Dev. 10: 81–93
Maitra A, Wistuba I, Washington C, Virmani A, Asfaq R, Milchgrub S, Gazdar A, Minna J . 2001 Am. J. Pathol. 159: 119–130
Mansour SL, Thomas KR, Capecci MR . 1988 Nature 336: 348–352
Mitelman F . 1991 Catalogue of chromosome aberrations in cancer New York: John Wiley
Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, Morgan NV, Moch H, Richards FM, Kishida T, Yao M, Schraml P, Latif F, Maher ER . 2001 Cancer Res. 61: 7277–7281
Ramirez-Solis R, Liu P, Bradley A . 1995 Nature 378: 720–724
Sekido Y, Ahmadian M, Wistuba I, Latif F, Bader S, Wei M-H, Duh F-M, Gazdar A, Lerman M, Minna JD . 1998 Oncogene 16: 3151–3157
Smith AJH, De Sousa MA, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts P . 1995 Nat. Genet. 9: 376–385
Thomas JW, LaMantia C, Magnuson T . 1998 Proc. Natl. Acad. Sci. USA 95: 1114–1119
Vogelstein B, Kinzler KW . 1993 Trends Genet. 9: 138–141
Wistuba I, Behrens C, Virmani A, Mele G, Milchgrub S, Girard L, Fondon JR, Garner H, McKay B, Latif F, Lerman MI, Lam S, Gazdar AF, Minna JD . 2000 Cancer Res. 60: 1949–1960
Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A . 2000 Mol. Cell. Biol. 20: 648–655
Zhu Y, Jong MC, Frazer KA, Gong E, Krauss RM, Cheng J-F, Boffelli D, Rubin EM . 2000 Proc. Natl. Acad. Sci. USA 97: 1137–1142
Acknowledgements
We thank Ms Lorraine Dobbie and Ms Diana Peddie for chimera generation, mouse breeding and genotyping; Dr Terence Rabbitts for the 129/Ola mouse lambda library; Drs Michael Lerman and Eric Lander for the BAC clone and Dr Lerman for human cDNA Luca clones. This work was funded by the award of a BBSRC project grant to AJH Smith (15/G05898) and an MRC programme grant to PH Rabbitts (G9703123).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Smith, A., Xian, J., Richardson, M. et al. Cre-loxP chromosome engineering of a targeted deletion in the mouse corresponding to the 3p21.3 region of homozygous loss in human tumours. Oncogene 21, 4521–4529 (2002). https://doi.org/10.1038/sj.onc.1205530
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1205530
Keywords
This article is cited by
-
Lineage tracing: technology tool for exploring the development, regeneration, and disease of the digestive system
Stem Cell Research & Therapy (2020)
-
Genetically modified mouse models in cancer studies
Clinical and Translational Oncology (2008)
-
Maximizing mouse cancer models
Nature Reviews Cancer (2007)