Key Points
-
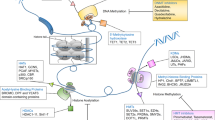
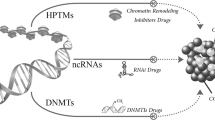
The past decade has seen a rapid emergence of epigenetics as a major contributor to carcinogenesis. Aberrations in the normal DNA methylation patterns and histone modifications have been recognized as targets for therapy which, unlike conventional chemotherapy, aim to revert the abnormal state of malignant cells to a more normal condition.
-
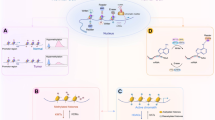
Demethylating agents that belong to a group of nucleoside analogues all have cytosine-ring modifications that allow each compound to form a covalent complex with a DNA methyltransferase, thereby inhibiting further methylation. Other DNA-methylation inhibitors belong to a group of non-nucleoside analogues whose mechanism of inhibition is not well known.
-
DNA methylation inhibitors have the disadvantages of lacking specificity and causing genome-wide hypomethylation which might activate appropriately silent genes and/or initiate genome instability, leading to undesirable consequences. These problems will be circumvented with more specific drugs directed to specific regions of the genome. It is anticipated that these drugs would be available in the form of chemically synthesized small molecules, which are more effective than cytidine analogues because they do not require incorporation into DNA and bind directly to the catalytic site of the DNA methyl transferases.
-
Histone deacetylase (HDAC) inhibitors are divided into four groups but hybrid molecules combining functional groups with superior inhibitory effects have already been synthesized. Compounds that inhibit individual members of all HDAC classes will be synthesized in the future.
-
Lysine methylation is another histone modification which could be essential in regulating gene expression but its use as a target for epigenetic therapy might not come to fruition until more complete classification of this type of epigenetic regulation is possible.
Abstract
The initiation and progression of cancer is controlled by both genetic and epigenetic events. Unlike genetic alterations, which are almost impossible to reverse, epigenetic aberrations are potentially reversible, allowing the malignant cell population to revert to a more normal state. With the advent of numerous drugs that target specific enzymes involved in the epigenetic regulation of gene expression, the utilization of epigenetic targets is emerging as an effective and valuable approach to chemotherapy as well as chemoprevention of cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). This is an excellent review on DNA methylation.
Takai, D. & Jones, P. A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA 99, 3740–3745 (2002).
Fujita, N. et al. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol. Cell Biol. 19, 6415–6426 (1999).
Hendrich, B. et al. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome 10, 906–912 (1999).
Perini, G., Diolaiti, D., Porro, A. & Della Valle, G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl Acad. Sci. USA 102, 12117–12122 (2005).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001). This review established the major impact histones have on chromatin as it relates to the regulation of gene expression.
Margueron, R., Trojer, P. & Reinberg, D. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15, 163–176 (2005).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). The first paper to show an inactive marker in the form of lysine methylation.
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Rice, J. C. et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 (2003).
Vakoc, C. R., Mandat, S. A., Olenchock, B. A. & Blobel, G. A. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 (2005).
Hashimshony, T., Zhang, J., Keshet, I., Bustin, M. & Cedar, H. The role of DNA methylation in setting up chromatin structure during development. Nature Genet. 34, 187–192 (2003).
Chen, W. et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell 6, 387–398 (2004).
Hoffmann, M. J. & Schulz, W. A. Causes and consequences of DNA hypomethylation in human cancer. Biochem. Cell Biol. 83, 296–321 (2005).
Holst, C. R. et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 63, 1596–1601 (2003).
Cheng, J. C. et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl Cancer Inst. 95, 399–409 (2003).
Herranz, M. et al. The novel DNA methylation inhibitor zebularine is effective against the development of murine T-cell lymphoma. Blood 20 Oct 2005 [epub ahead of print].
Laird, P. W. et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81, 197–205 (1995). This is an important work which demonstrated that DNMT1 activity is closely linked to tumorigenesis and that the enzyme may be targeted for treatment of cancer.
Feinberg, A. P. & Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983).
Riggs, A. D. & Jones, P. A. 5-methylcytosine, gene regulation, and cancer. Adv. Cancer Res. 40, 1–30 (1983).
Eden, A., Gaudet, F., Waghmare, A. & Jaenisch, R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455 (2003).
Gaudet, F. et al. Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003). A discussion of the impact of genomic hypomethylation of DNA on the formation of cancer.
Gius, D. et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 6, 361–371 (2004).
Cheng, J. C. et al. Preferential response of cancer cells to zebularine. Cancer Cell 6, 151–158 (2004).
Liang, G., Gonzales, F. A., Jones, P. A., Orntoft, T. F. & Thykjaer, T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 62, 961–966 (2002).
Costello, J. F. et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet. 24, 132–138 (2000). This paper demonstrated that abnormal DNA methylation patterns exist in cancer.
Toyota, M. et al. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA 96, 8681–8686 (1999). This paper showed the presence of the CpG island methylator phenotype in cancer.
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genet. 37, 853–862 (2005).
Fraga, M. F. et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genet. 37, 391–400 (2005).
Seligson, D. B. et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435, 1262–1266 (2005).
Mutskov, V. & Felsenfeld, G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 23, 138–149 (2004).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Whetstine, J. R. et al. Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol. Cell 18, 483–490 (2005).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). The first identification of the histone demethylase enzyme.
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature (2005).
Garcia-Cao, M., O'Sullivan, R., Peters, A. H., Jenuwein, T. & Blasco, M. A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nature Genet. 36, 94–99 (2004).
Erlanson, D. A., Chen, L., and Verdine, G. L. DNA Methylation through a locally unpaired intermediate. J. Am. Chem. Soc. 115, 12583–12584 (1993).
Klimasauskas, S., Kumar, S., Roberts, R. J. & Cheng, X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 (1994). This paper marks the elucidation of molecular mechanism of methyltransferase enzyme by X-ray crystallography.
Wu, J. C. & Santi, D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 262, 4778–4786 (1987).
Santi, D. V., Garrett, C. E. & Barr, P. J. On the mechanism of inhibition of DNA-cytosine methyl-transferases by cytosine analogs. Cell 33, 9–10 (1983).
Santi, D. V., Norment, A. & Garrett, C. E. Covalent bond formation between a DNA-cytosine methyl-transferase and DNA containing 5-azacytosine. Proc. Natl Acad. Sci. USA 81, 6993–6997 (1984).
Zhou, L. et al. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321, 591–599 (2002).
Ghoshal, K. et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell Biol. 25, 4727–4741 (2005).
Schermelleh, L. et al. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nature Methods 2, 751–756 (2005).
Pliml, J. & Sorm, F. Synthesis of 2′-deoxy-D-ribofuranosyl-5-azacytosine. Coll Czech Chem Commun 29, 2576–2577 (1964).
Sorm, F. & Vesely, J. Effect of 5-aza-2′-deoxycytidine against leukemic and hematopoietic tissues in AKR mice. Neoplasma 15, 339–343 (1968).
Jones, P. A. & Taylor, S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980).
Taylor, S. M. & Jones, P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17, 771–779 (1979). References 47 and 48 showed for the first time that DNA methylation is inhibited in mammalian cells by the cytidine analogues 5-azacytidine and 5-aza-2′-deoxycytidine.
Lin, K. T., Momparler, R. L. & Rivard, G. E. High-performance liquid chromatographic analysis of chemical stability of 5-aza-2′-deoxycytidine. J. Pharm. Sci. 70, 1228–1232 (1981).
Notari, R. E. & DeYoung, J. L. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J. Pharm. Sci. 64, 1148–1157 (1975).
Kaminskas, E. et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin. Cancer Res. 11, 3604–3608 (2005).
Issa, J. P. et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103, 1635–1640 (2004).
Issa, J. P. et al. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J. Clin. Oncol. 23, 3948–3956 (2005).
Lubbert, M. et al. Nonclonal neutrophil responses after successful treatment of myelodysplasia with low-dose 5-aza-2′-deoxycytidine (decitabine). Leuk. Res. 28, 1267–1271 (2004). References 52–54 showed that a low-dose regimen of decitabine was more efficacious than high-dose treatment in observing demethylation in patients.
Aparicio, A. et al. Phase I trial of continuous infusion 5-aza-2′-deoxycytidine. Cancer Chemother. Pharmacol. 51, 231–239 (2003).
Cameron, E. E., Bachman, K. E., Myohanen, S., Herman, J. G. & Baylin, S. B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet. 21, 103–107 (1999).
Rudek, M. A. et al. Pharmacokinetics of 5-azacitidine administered with phenylbutyrate in patients with refractory solid tumors or hematologic malignancies. J. Clin. Oncol. 23, 3906–3911 (2005).
Beisler, J. A., Abbasi, M. M. & Driscoll, J. S. Dihydro-5-azacytidine hydrochloride, a biologically active and chemically stable analog of 5-azacytidine. Cancer Treat. Rep. 60, 1671–1674 (1976).
Beisler, J. A., Abbasi, M. M., Kelley, J. A. & Driscoll, J. S. Synthesis and antitumor activity of dihydro-5-azacytidine, a hydrolytically stable analogue of 5-azacytidine. J. Med. Chem. 20, 806–812 (1977).
Presant, C. A., Coulter, D., Valeriote, F. & Vietti, T. J. Contrasting cytotoxicity kinetics of 5-azacytidine and dihydro-5-azacytidine hydrochloride in L1210 leukemia in mice. J. Natl Cancer Inst. 66, 1151–1154 (1981).
Stopper, H., Korber, C., Gibis, P., Spencer, D. L. & Caspary, W. J. Micronuclei induced by modulators of methylation: analogs of 5-azacytidine. Carcinogenesis 16, 1647–1650 (1995).
Antonsson, B. E., Avramis, V. I., Nyce, J. & Holcenberg, J. S. Effect of 5-azacytidine and congeners on DNA methylation and expression of deoxycytidine kinase in the human lymphoid cell lines CCRF/CEM/0 and CCRF/CEM/dCk-1. Cancer Res. 47, 3672–3678 (1987).
Kees, U. R. & Avramis, V. I. Biochemical pharmacology and DNA methylation studies of arabinosyl 5-azacytidine and 5, 6-dihydro-5-azacytidine in two human leukemia cell lines PER-145 and PER-163. Anticancer Drugs 6, 303–310 (1995).
Powell, W. C. & Avramis, V. I. Biochemical pharmacology of 5, 6-dihydro-5-azacytidine (DHAC) and DNA hypomethylation in tumor (L1210)-bearing mice. Cancer Chemother. Pharmacol. 21, 117–121 (1988).
Curt, G. A. et al. A phase I and pharmacokinetic study of dihydro-5-azacytidine (NSC 264880). Cancer Res. 45, 3359–3363 (1985).
Eidinoff, M. L., Rich, M. A. & Perez, A. G. Growth inhibition of a human tumor cell strain by 5-fluorocytidine and 5-fluoro-2′-deoxycytidine: reversal studies. Cancer Res. 19, 638–642 (1959).
Chen, L. et al. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30, 11018–11025 (1991).
Barchi, J. J., Jr. et al. Improved synthesis of zebularine [1-(β-D-ribofuranosyl)-dihydropyrimidin-2-one] nucleotides as inhibitors of human deoxycytidylate deaminase. J. Enzyme Inhib. 9, 147–162 (1995).
Frick, L., Yang, C., Marquez, V. E. & Wolfenden, R. Binding of pyrimidin-2-one ribonucleoside by cytidine deaminase as the transition-state analogue 3,4-dihydrouridine and the contribution of the 4-hydroxyl group to its binding affinity. Biochemistry 28, 9423–9430 (1989).
Kim, C. H., Marquez, V. E., Mao, D. T., Haines, D. R. & McCormack, J. J. Synthesis of pyrimidin-2-one nucleosides as acid-stable inhibitors of cytidine deaminase. J. Med. Chem. 29, 1374–1380 (1986).
Laliberte, J., Marquez, V. E. & Momparler, R. L. Potent inhibitors for the deamination of cytosine arabinoside and 5-aza-2′-deoxycytidine by human cytidine deaminase. Cancer Chemother. Pharmacol. 30, 7–11 (1992).
Driscoll, J. S. et al. Antitumor properties of 2(1H)-pyrimidinone riboside (zebularine) and its fluorinated analogues. J. Med. Chem. 34, 3280–3284 (1991).
Lee, G., Wolff, E. & Miller, J. H. Mutagenicity of the cytidine analog zebularine in Escherichia coli. DNA Repair (Amst) 3, 155–161 (2004).
Holleran, J. L. et al. Plasma pharmacokinetics, oral bioavailability, and interspecies scaling of the DNA methyltransferase inhibitor, zebularine. Clin. Cancer Res. 11, 3862–3868 (2005).
Brueckner, B. et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 65, 6305–6311 (2005).
Schuebel, K. & Baylin, S. In living color: DNA methyltransferase caught in the act. Nature Methods 2, 736–738 (2005).
Davis, A. J. et al. Phase I and pharmacologic study of the human DNA methyltransferase antisense oligodeoxynucleotide MG98 given as a 21-day continuous infusion every 4 weeks. Invest. New Drugs 21, 85–97 (2003).
Stewart, D. J. et al. A phase I pharmacokinetic and pharmacodynamic study of the DNA methyltransferase 1 inhibitor MG98 administered twice weekly. Ann. Oncol. 14, 766–774 (2003).
Pina, I. C. et al. Psammaplins from the sponge Pseudoceratina purpurea: inhibition of both histone deacetylase and DNA methyltransferase. J. Org. Chem. 68, 3866–3873 (2003).
Fang, M. Z. et al. Tea polyphenol (–)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 63, 7563–7570 (2003).
Alikhani-Koopaei, R., Fouladkou, F., Frey, F. J. & Frey, B. M. Epigenetic regulation of 11 β-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Invest. 114, 1146–1157 (2004).
Cornacchia, E. et al. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 140, 2197–2200 (1988).
Lin, X. et al. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 61, 8611–8616 (2001).
Segura-Pacheco, B. et al. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res. 9, 1596–1603 (2003).
Villar-Garea, A., Fraga, M. F., Espada, J. & Esteller, M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res 63, 4984–4989 (2003).
Zambrano, P. et al. A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer 5, 44 (2005).
Chuang, J. C. et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol. Cancer Ther. 4, 1515–1520 (2005).
Rocchi, P. et al. p21Waf1/Cip1 is a common target induced by short-chain fatty acid HDAC inhibitors (valproic acid, tributyrin and sodium butyrate) in neuroblastoma cells. Oncol. Rep. 13, 1139–1144 (2005).
Nebbioso, A. et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nature Med. 11, 77–84 (2005).
Peart, M. J. et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA 102, 3697–3702 (2005).
Shetty, S. et al. Transcription factor NF-κB differentially regulates death receptor 5 expression involving histone deacetylase 1. Mol. Cell Biol. 25, 5404–5416 (2005).
Dai, Y., Rahmani, M., Dent, P. & Grant, S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-κB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol. Cell Biol. 25, 5429–5444 (2005).
Duan, H., Heckman, C. A. & BOX er, L. M. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell Biol. 25, 1608–1619 (2005).
Michaelis, M. et al. Valproic acid inhibits angiogenesis in vitro and in vivo. Mol. Pharmacol. 65, 520–527 (2004).
Joseph, J. et al. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene 23, 6304–6315 (2004).
Qiu, L. et al. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol. Biol Cell. 11, 2069–2083 (2000).
Gu, W. & Roeder, R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Chen, L. F. & Greene, W. C. Regulation of distinct biological activities of the NF-κB transcription factor complex by acetylation. J. Mol. Med. 81, 549–557 (2003).
Stadtman, E. R. & Barker, H. A. Fatty acid synthesis by enzyme preparations of Clostridium kluyveri; a consideration of postulated 4-carbon intermediates in butyrate synthesis. J. Biol. Chem. 181, 221–235 (1949).
Gottlicher, M. et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20, 6969–6978 (2001).
Candido, E. P., Reeves, R. & Davie, J. R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14, 105–113 (1978).
Sealy, L. & Chalkley, R. The effect of sodium butyrate on histone modification. Cell 14, 115–121 (1978).
Raffoux, E., Chaibi, P., Dombret, H. & Degos, L. Valproic acid and all-trans retinoic acid for the treatment of elderly patients with acute myeloid leukemia. Haematologica 90, 986–988 (2005).
Yang, H., Hoshino, K., Sanchez-Gonzalez, B., Kantarjian, H. & Garcia-Manero, G. Antileukemia activity of the combination of 5-aza-2′-deoxycytidine with valproic acid. Leuk. Res. 29, 739–748 (2005).
Kramer, O. H. et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22, 3411–3420 (2003).
Perrine, S. P. et al. Butyrate derivatives. New agents for stimulating fetal globin production in the β-globin disorders. Am. J. Pediatr. Hematol. Oncol. 16, 67–71 (1994).
van de Mark, K., Chen, J. S., Steliou, K., Perrine, S. P. & Faller, D. V. Alpha-lipoic acid induces p27Kip-dependent cell cycle arrest in non-transformed cell lines and apoptosis in tumor cell lines. J. Cell. Physiol. 194, 325–340 (2003).
Yoshida, M., Kijima, M., Akita, M. & Beppu, T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179 (1990).
Kelly, W. K. et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin. Cancer Res. 9, 3578–3588 (2003).
Kelly, W. K. et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J. Clin. Oncol. 23, 3923–3931 (2005).
Finnin, M. S. et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 (1999).
Monneret, C. Histone deacetylase inhibitors. Eur J. Med. Chem. 40, 1–13 (2005).
Giles, F. J. et al. A phase I/II study of intravenous LBH589, a novel histone deacetylase (HDAC) inhibitor, in patients (pts) with advanced hematologic malignancies. Blood 104, 499A (2004).
Remiszewski, S. W. et al. N-hydroxy-3-phenyl-2-propenamides as novel inhibitors of human histone deacetylase with in vivo antitumor activity: discovery of (2E)-N-hydroxy-3-[4-[[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl]phenyl]-2-propenamide (NVP-LAQ824). J. Med. Chem. 46, 4609–4624 (2003).
Kijima, M., Yoshida, M., Sugita, K., Horinouchi, S. & Beppu, T. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J. Biol. Chem. 268, 22429–22435 (1993).
Byrd, J. C. et al. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 105, 959–967 (2005).
Marshall, J. L. et al. A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J. Exp. Ther. Oncol. 2, 325–332 (2002).
Piekarz, R. L. et al. Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood 98, 2865–2868 (2001).
Sandor, V. et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin. Cancer Res. 8, 718–728 (2002).
Remiszewski, S. W. The discovery of NVP-LAQ824: from concept to clinic. Curr. Med. Chem. 10, 2393–2402 (2003).
Singh, S. B. et al. Structure, histone deacetylase, and antiprotozoal activities of apicidins B and C, congeners of apicidin with proline and valine substitutions. Org. Lett. 3, 2815–2818 (2001).
Furumai, R. et al. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl Acad. Sci. USA 98, 87–92 (2001).
Jose, B. et al. Novel histone deacetylase inhibitors: cyclic tetrapeptide with trifluoromethyl and pentafluoroethyl ketones. Bioorg. Med. Chem. Lett. 14, 5343–5346 (2004).
Nishino, N. et al. Cyclic tetrapeptides bearing a sulfhydryl group potently inhibit histone deacetylases. Org. Lett. 5, 5079–5082 (2003).
Nishino, N. et al. Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg. Med. Chem. Lett. 14, 2427–2431 (2004).
Saito, A. et al. A synthetic inhibitor of histone deacetylase, MS-27–275, with marked in vivo antitumor activity against human tumors. Proc. Natl Acad. Sci. USA 96, 4592–4597 (1999).
Camphausen, K. et al. Enhanced radiation-induced cell killing and prolongation of gammaH2AX foci expression by the histone deacetylase inhibitor MS-275. Cancer Res. 64, 316–321 (2004).
Wang, X. F. et al. Epigenetic modulation of retinoic acid receptor β2 by the histone deacetylase inhibitor MS-275 in human renal cell carcinoma. Clin. Cancer Res. 11, 3535–3542 (2005).
Pauer, L. R. et al. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer Invest. 22, 886–896 (2004).
Prakash, S. et al. Chronic oral administration of CI-994: a phase 1 study. Invest. New Drugs 19, 1–11 (2001).
Undevia, S. D. et al. A phase I study of the oral combination of CI-994, a putative histone deacetylase inhibitor, and capecitabine. Ann. Oncol. 15, 1705–1711 (2004).
Yoo, C. B., Cheng, J. C. & Jones, P. A. Zebularine: a new drug for epigenetic therapy. Biochem. Soc. Trans. 32, 910–912 (2004).
Pohlmann, P. et al. Phase II trial of cisplatin plus decitabine, a new DNA hypomethylating agent, in patients with advanced squamous cell carcinoma of the cervix. Am J. Clin. Oncol. 25, 496–501 (2002).
Schwartsmann, G. et al. A phase I trial of cisplatin plus decitabine, a new DNA-hypomethylating agent, in patients with advanced solid tumors and a follow-up early phase II evaluation in patients with inoperable non-small cell lung cancer. Invest. New Drugs 18, 83–91 (2000).
Laird, P. W. The power and the promise of DNA methyl-ation markers. Nature Rev. Cancer 3, 253–266 (2003).
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 19 Nov 2004 [epub ahead of print].
Kubicek, S. & Jenuwein, T. A crack in histone lysine methylation. Cell 119, 903–906 (2004).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000). An important paper that makes a connection between histone methylation and HP1.
Liang, G. et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA 101, 7357–7362 (2004).
Schubeler, D. et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 (2004).
Lo, W. S. et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926 (2000).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bourc'his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004).
Bourc'his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Gibbons, R. J. Histone modifying and chromatin remodelling enzymes in cancer and dysplastic syndromes. Hum. Mol. Genet. 14 (Spec. No 1), R85–R92 (2005).
Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C. & Horikawa, I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Mol. Biol. Cell. 16, 4623–4635 (2005).
Schneider, R., Bannister, A. J. & Kouzarides, T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 27, 396–402 (2002).
Kouzarides, T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12, 198–209 (2002).
Bannister, A. J. & Kouzarides, T. Reversing histone methylation. Nature 436, 1103–1106 (2005).
Karon, M. et al. 5-Azacytidine: a new active agent for the treatment of acute leukemia. Blood 42, 359–365 (1973).
Moertel, C. G., Schutt, A. J., Reitemeier, R. J. & Hahn, R. G. Phase II study of 5-azacytidine (NSC-102816) in the treatment of advanced gastrointestinal cancer. Cancer Chemother. Rep. 56, 649–652 (1972).
Gaynon, P. S. & Baum, E. S. Continuous infusion of 5-azacytidine as induction for acute nonlymphocytic leukemia in patients with previous exposure to 5-azacytidine. Oncology 40, 192–194 (1983).
Velez-Garcia, E., Vogler, W. R., Bartolucci, A. A. & Arkun, S. N. Twice weekly 5-azacytidine infusion in dissmeinated metastatic cancer: a phase II study. Cancer Treat. Rep. 61, 1675–1677 (1977).
Weiss, A. J. et al. Phase II study of 5-azacytidine in solid tumors. Cancer Treat. Rep. 61, 55–58 (1977).
Lomen, P. L., Khilanani, P. & Kessel, D. Phase I study using combination of hydroxyurea and 5-azacytidine (NSC-102816). Neoplasma 27, 101–106 (1980).
Nitschke, R., Land, V., Steuber, C. P. & Ragab, A. H. Low response rate to 5 aza-cytidine, vincristine, and prednisone therapy in previously treated childhood acute nonlymphocytic leukemia: a Southwest Oncology Group Study. Am. J. Pediatr. Hematol. Oncol. 3, 307–309 (1981).
Kornblith, A. B. et al. Impact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized phase III trial: a Cancer and Leukemia Group B study. J. Clin. Oncol. 20, 2441–2452 (2002).
Silverman, L. R. et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 20, 2429–2440 (2002).
Silverman, L. R. et al. Effects of treatment with 5-azacytidine on the in vivo and in vitro hematopoiesis in patients with myelodysplastic syndromes. Leukemia 7 (Suppl. 1), 21–29 (1993).
Momparler, R. L. et al. Pilot phase I-II study on 5-aza-2′-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anticancer Drugs 8, 358–368 (1997).
Zagonel, V. et al. 5-Aza-2′-deoxycytidine (Decitabine) induces trilineage response in unfavourable myelodysplastic syndromes. Leukemia 7 (Suppl. 1), 30–35 (1993).
Samlowski, W. E. et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: inhibition of promoter-specific and global genomic DNA methylation. J. Clin. Oncol. 23, 3897–3905 (2005).
Sarraf, S. A. & Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 (2004).
Zgouras, D., Becker, U., Loitsch, S. & Stein, J. Modulation of angiogenesis-related protein synthesis by valproic acid. Biochem. Biophys. Res. Commun. 316, 693–697 (2004).
Acknowledgements
We are grateful to J. Lin for help with the drawing of a figure and G. Egger, A. Aparicio and A. Yang for proofreading of the manuscript and their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
P.A.J. is a consultant to and shareholder of Epigenomics AG and is on the Speakers Bureau for MGI Pharma which is developing decitabine. C.B.Y. does not have any competing financial interest.
Glossary
- Epigenetics
-
Mitotically and meiotically heritable changes in gene expression patterns that are not explained by DNA sequence changes. Epigenetic changes are implicated in many aspects of cell biology including X-inactivation, imprinting, position-effect variegation and, most recently, cancer.
- DNA methylation
-
A modification to the 5-position of a cytosine ring that occurs most commonly in the context of a CpG palindrome in DNA that can be transmitted to daughter cells at cell division and has been implicated in regulation of gene expression.
- CpG island
-
A region of DNA that is about 500-bp long with a high GC content and the ratio of observed to expected CpG dinucleotides greater than 0.6, which usually lies near the promoter of a gene and that is unmethylated in the germline.
- Histone methylation
-
Histone methylation on lysine and arginine residues is mediated by histone lysine methyltransferases (HMTs) and protein arginine methyltransferases (PRMTs), respectively, which transfer a methyl group from S-adenosyl-L-methionine. Lysine residues are mono-, di- or trimethylated and arginines are mono-, symmetric di- or asymmetric dimethylated on their ε-amino groups, and these methyl marks play an important role in chromatin remodelling.
- Histone lysine acetylation
-
Histones are acetylated on the ε-amino group of lysine residues. Acetyl groups interact with the bromodomain of other proteins, and acetylation of H3 and H4 is implicated in transcriptional activation.
- SET domain
-
SET (Su(var)39, Enhancer of zeste, Trithorax) domains are protein–protein interacting domains that are associated with methyltransferase activity and function to modulate the chromatin structure.
- Restriction landmark genomic scanning
-
(RLGS). A quantitative method for measuring the level of DNA methylation in thousands of CpG islands. This technique is especially suited for detection of aberrant DNA methylation and DNA amplifications.
- Androgen receptor
-
(AR). A member of a family of proteins called nuclear receptors which bind lipophilic steroid molecules. AR interacts with androgens, binds to androgen receptor element (ARE), and causes transcription activation of its target genes; it is a common target of therapy for prostate cancer.
- Uridine/cytidine kinase
-
An enzyme that catalyses the phosphorylation of uridine and cytidine into uridylate and cytidylate, respectively. It also phosphorylates 5-azacytidine and zebularine into their respective monophosphate moieties.
- S-adenosyl-L-methionine
-
An enzymatic cofactor and the most important methyl donor in various biological systems. It is involved in both DNA and histone methylation.
- Myelodysplasia
-
A characteristic of many haematological conditions that occurs as a result of abnormal bone marrow function with a likelihood of progressing into acute myelogenous leukaemia.
- Epigenome
-
The comprehensive collection of genome-wide DNA-methylation patterns and chromatin modifications, which gives structure and function to the genome.
Rights and permissions
About this article
Cite this article
Yoo, C., Jones, P. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5, 37–50 (2006). https://doi.org/10.1038/nrd1930
Issue Date:
DOI: https://doi.org/10.1038/nrd1930
This article is cited by
-
DNA methylation in the genesis, progress and prognosis of head and neck cancer
Holistic Integrative Oncology (2023)
-
Fetal hemoglobin level predicts lower-risk myelodysplastic syndrome
International Journal of Hematology (2023)
-
BRD4770 inhibits vascular smooth muscle cell proliferation via SUV39H2, but not EHMT2 to protect against neointima formation
Human Cell (2023)
-
Evaluation of antibacterial, antioxidant, cytotoxic, and acetylcholinesterase inhibition activities of novel [1,4] benzoxazepines fused to heterocyclic systems with a molecular modeling study
Medicinal Chemistry Research (2023)
-
Unique combinations of epigenetic modifiers synergistically impair the viability of the U87 glioblastoma cell line while exhibiting minor or moderate effects on normal stem cell growth
Medical Oncology (2022)