Abstract
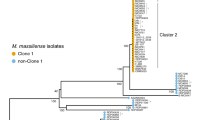
The rapid increase in Pseudomonas (Burkholderia) cepacia infection in cystic fibrosis (CF) patients suggests epidemic transmission, but the degree of transmissibility remains controversial as conflicting conclusions have been drawn from studies at different CF centres. This report provides the first DNA sequence-based documentation of a divergent evolutionary lineage of P. cepacia associated with CF centre epidemics in North America (Toronto) and Europe (Edinburgh). The involved epidemic clone encoded and expressed novel cable (Cbl) pili that bind to CF mucin. The sequence of the cbIA pilin subunit gene carried by the epidemic isolates proved to be invariant. Although it remains to be determined how many distinct, highly transmissible lineages exist, our results provide both a DNA sequence and chromosomal fingerprint that can be used to screen for one such particularly infectious, transatlantic clone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
LiPuma, J., Mortensen, J., Dasen, S. & Stull, T. Ribotype analysis of Pcepacia forn cystic fibrosis treatment centers. J. Pediat 113, 859–862 (1988).
LiPuma, J., Dasen, S., Nielson, D., Stern, R. & Stull, T. Person-to-person transmission of P. cepacia between patients with cystic fibrosis. Lancet 336, 1094–1096 (1990).
Govan, J.R.W. et al. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342, 15–19 (1993).
Sajjan, U., Sun, L., Goldstein, R. & Forstner, J. Cable (Cbl) type II pili of cystic fi-brosis-associated Burkholderia (Pseudomonas) cepacia: Nucleotide sequence of the cbIA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacterial. 177, 1030–1038 (1995).
Tablan, O. et al. Pseudomonas cepacia colonization in patients with cystic fibrosis: Risk factors and clinical outcome. J. Pediat. 107, 382–387 (1985).
Thomassen, M.J., Demko, C., Klinger, J. & Stern, R. Pseudomonas cepacia colonization among patients with cystic fibrosis: A new opportunist. Am. Rev. respir. Dis. 131, 791–796 (1985).
Ramirez, J.C. et al. Bilateral lung transplantation for cystic fibrosis. J. thor. cardiovasc. Surg. 103, 287–294 (1992).
Snell, G.I., deHoyos, A., Krajden, M., Winton, T. & Maurer, J.R. Pseudomonas cepacia in lung transplant recipients with cystic fibrosis. Chest 103, 466–471 (1993).
Steinbach, S. et al. Transmissibility of Pseudomonas cepacia infection in clinic patients and lung-transplant recipients with cystic fibrosis. New Engl. J. Med. 331, 981–987 (1994).
Anderson, D.J., Kuhns, J.S., Vasil, M.L., Gerding, D.N. & Janoff, E.N. DNA fingerprinting by pulsed field gel electrophoresis and ribotyping to distinguish Pseudomonas cepacia isolates from a nosocomial outbreak. J. clin. Microbiol. 29, 648–649 (1991).
Sajjan, U., Corey, M., Karmali, M. & Forstner, J. Binding of Pseudomonas cepacia to normal human intestinal mucin and respiratory mucin from patients with cystic fibrosis. J. clin. Invest 89, 648–556 (1992).
Sajjan, U. & Forstner, J. Role of a 22-kilodalton pilin protein in binding of Pseudomonas cepacia to buccal epithelial cells. Infect. Immun. 61, 3156–3163 (1993).
Goldstein, R. et al. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia {Pseudomonas) cepacia . J. Bacteriol. 177, 1039–1052 (1995).
Dice, L. Measures of the amount of ecological association between species. Ecology 26, 297–302 (1945).
Saitou, N. & Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molec. biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985).
Nelson, K. & Selander, R. Evolutionary genetics of the proline permease (putP) and the control region of the proline utilization operon in populations of Salmonella and E. coli . J. Bacteriol. 174, 6886–6895 (1992).
Smith, N. & Selander, R. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium . J. Bacteriol 172, 603–609 (1990).
Thomassen, M.J., Demko, C., Doershuk, C., Stern, R.C. & Klinger, J. Pseudomonas cepacia: Decrease in colonization in patients with cystic fibrosis. Am. Rev. respir. Dis. 134, 669–671 (1986).
Vinograd, J. & Hearst, J.E. Equilibrium sedimentation of macromolecules and viruses in a density gradient. Fortsch. Chem. Organisch. Naturstoffe. 20, 372–422 (1962).
Mullis, K. & Faloona, F. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Meth. Enzym. 155, 335–350 (1987).
Sanger, F., Nicklen, S. & Coulson, A. DNA sequencing with chain-terminating inhibitors. Proc. natn. Acad. Sci. U.S.A. 74, 5463–5467 (1977).
Prevost, G., Jaulhac, B. & Piemont, Y. DNA fingerprinting by PFGE is more effective than ribotyping in distinguishing among MRSA isolates. J. clin. Microbiol. 30, 967–973 (1992).
Rabkin, C., Jams, W., Anderson, R., Stull, T. & Woods, D. P. cepacia typing systems: Collaborative study to assess their potential in epidemiologic investigations. Rev. infect. Dis. 11, 600–607 (1989).
Nei, M. & Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. natn. Acad. Sci. U.S.A. 76, 5269–5273 (1979).
Arthur, M. et al. Restriction fragment length polymorphisms among uropatho-genic Escherichia coli: pop-related sequences compared with rrn operons. Infect. Immun. 58, 471–479 (1990).
Arthur, M. et al. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli . Infect. Immun. 57, 303–313 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sun, L., Jiang, RZ., Steinbach, S. et al. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat Med 1, 661–666 (1995). https://doi.org/10.1038/nm0795-661
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nm0795-661
This article is cited by
-
Lung transplantation: infection, inflammation, and the microbiome
Seminars in Immunopathology (2011)
-
Bovine lactoferrin interacts with cable pili of Burkholderia cenocepacia
BioMetals (2010)
-
Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients
European Journal of Clinical Microbiology & Infectious Diseases (2009)
-
Development and application of the active surveillance of pathogens microarray to monitor bacterial gene flux
BMC Microbiology (2008)
-
Clinical and Environmental Burkholderia Strains: Biofilm Production and Intracellular Survival
Current Microbiology (2007)