Abstract
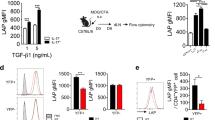
Here we describe a reporter mouse strain designed to map the fate of cells that have activated interleukin 17A (IL-17A). We found that IL-17-producing helper T cells (TH17 cells) had distinct plasticity in different inflammatory settings. Chronic inflammatory conditions in experimental autoimmune encephalomyelitis (EAE) caused a switch to alternative cytokines in TH17 cells, whereas acute cutaneous infection with Candida albicans did not result in the deviation of TH17 cells to the production of alternative cytokines, although IL-17A production was shut off in the course of the infection. During the development of EAE, interferon-γ (IFN-γ) and other proinflammatory cytokines in the spinal cord were produced almost exclusively by cells that had produced IL-17 before their conversion by IL-23 ('ex-TH17 cells'). Thus, this model allows the actual functional fate of effector T cells to be related to TH17 developmental origin regardless of IL-17 expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Harrington, L.E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005).
Park, H. et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 (2005).
Veldhoen, M. et al. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Ivanov, I.I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Yang, X.O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28, 29–39 (2008).
Annunziato, F. et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204, 1849–1861 (2007).
Bending, D. et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 119, 565–572 (2009).
Lee, Y.K. et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009).
Martin-Orozco, N. et al. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur. J. Immunol. 39, 216–224 (2009).
Shi, G. et al. Phenotype switching by inflammation-inducing polarized Th17 cells, but not by Th1 cells. J. Immunol. 181, 7205–7213 (2008).
Bettelli, E. & Kuchroo, V.K. IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together. J. Exp. Med. 201, 169–171 (2005).
Lexberg, M.H. et al. Th memory for interleukin-17 expression is stable in vivo. Eur. J. Immunol. 38, 2654–2664 (2008).
Hirota, K., Martin, B. & Veldhoen, M. Development, regulation and functional capacities of Th17 cells. Semin. Immunopathol. 32, 3–16 (2010).
Spits, H. & Di Santo, J.P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Luger, D. et al. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J. Exp. Med. 205, 799–810 (2008).
Bettelli, E. et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197, 1073–1081 (2003).
McGeachy, M.J. et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10, 314–324 (2009).
Zhou, L., Chong, M.M. & Littman, D.R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009).
de Beaucoudrey, L. et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J. Exp. Med. 205, 1543–1550 (2008).
Milner, J.D. et al. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776 (2008).
Kisand, K. et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J. Exp. Med. 207, 299–308 (2010).
O′Shea, J.J. & Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010).
Murphy, K.M. & Stockinger, B. Effector T cell plasticity: flexibility in the fact of changing circumstances. Nat. Immunol. 11, 674–680 (2010).
Mukasa, R. et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32, 616–627 (2010).
Wei, G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009).
Croxford, A.L., Kurschus, F.C. & Waisman, A. Cutting edge: an IL-17F-CreEYFP reporter mouse allows fate mapping of Th17 cells. J. Immunol. 182, 1237–1241 (2009).
Chang, S.H. & Dong, C. IL-17F: regulation, signaling and function in inflammation. Cytokine 46, 7–11 (2009).
Ahern, P.P. et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288 (2010).
Veldhoen, M., Hocking, R.J., Flavell, R.A. & Stockinger, B. Signals mediated by transforming growth factor-β initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7, 1151–1156 (2006).
Ponomarev, E.D. et al. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J. Immunol. 178, 39–48 (2007).
McQualter, J.L. et al. Granulocyte macrophage colony-stimulating factor: a new putative therapeutic target in multiple sclerosis. J. Exp. Med. 194, 873–882 (2001).
Boniface, K. et al. Human Th17 cells comprise heterogeneous subsets including IFN-γ-producing cells with distinct properties from the Th1 lineage. J. Immunol. 185, 679–687 (2010).
Stevens, E.A., Mezrich, J.D. & Bradfield, C.A. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology 127, 299–311 (2009).
Szabo, S.J., Dighe, A.S., Gubler, U. & Murphy, K.M. Regulation of the interleukin (IL)-12R β2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 185, 817–824 (1997).
Hegazy, A.N. et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 32, 116–128 (2010).
Bailey-Bucktrout, S.L. et al. Cutting edge: central nervous system plasmacytoid dendritic cells regulate the severity of relapsing experimental autoimmune encephalomyelitis. J. Immunol. 180, 6457–6461 (2008).
Isaksson, M. et al. Plasmacytoid DC promote priming of autoimmune Th17 cells and EAE. Eur. J. Immunol. 39, 2925–2935 (2009).
Shinohara, M.L., Kim, J.H., Garcia, V.A. & Cantor, H. Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: role of intracellular osteopontin. Immunity 29, 68–78 (2008).
Axtell, R.C. et al. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat. Med. 16, 406–412 (2010).
Stromnes, I.M. et al. Differential regulation of central nervous system autoimmunity by TH1 and TH17 cells. Nat. Med. 14, 337–342 (2008).
Lees, J.R. et al. Regional CNS responses to IFN-γ determine lesion localization patterns during EAE pathogenesis. J. Exp. Med. 205, 2633–2642 (2008).
Steinman, L. A rush to judgment on Th17. J. Exp. Med. 205, 1517–1522 (2008).
O'Connor, R.A. et al. Cutting edge: Th1 cells facilitate the entry of Th17 cells to the central nervous system during experimental autoimmune encephalomyelitis. J. Immunol. 181, 3750–3754 (2008).
McGeachy, M.J. et al. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nat. Immunol. 8, 1390–1397 (2007).
Suryani, S. & Sutton, I. An interferon-γ-producing Th1 subset is the major source of IL-17 in experimental autoimmune encephalitis. J. Neuroimmunol. 183, 96–103 (2007).
McGeachy, M.J. & Cua, D.J. Th17 cell differentiation: the long and winding road. Immunity 28, 445–453 (2008).
Michel, M.L. et al. Identification of an IL-17-producing NK1.1− iNKT cell population involved in airway neutrophilia. J. Exp. Med. 204, 995–1001 (2007).
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-beta signalling. Nature 467, 967–971 (2010).
Pepper, M. et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat. Immunol. 11, 83–89 (2010).
Sutton, C. et al. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203, 1685–1691 (2006).
Shimshek, D.R. et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26 (2002).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Acknowledgements
We thank J.-C. Renauld (Ludwig Institute for Cancer Research) for monoclonal antibody AM22.3; the Large Scale Facility of the Medical Research Council National Institute for Medical Research for growing C. albicans; G. Preece for cell sorting; and Biological Services (Medical Research Council National Institute for Medical Research) for breeding and maintenance of our mouse strains. Supported by the Medical Research Council UK (U117512792 to B.S. and U117563359 to A.J.P.) and the European Research Council (ref. 232782).
Author information
Authors and Affiliations
Contributions
K.H., J.H.D., H.A. and C.W. designed and did experiments; M.V., E.H., Y.L., M.T., U.M. and A.G. were involved in generating the construct, targeting embryonic stem cells and injections; D.J.C. provided IL-23p19-deficient mice; and A.J.P. and B.S. designed experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–2 and Supplementary Methods (PDF 2092 kb)
Rights and permissions
About this article
Cite this article
Hirota, K., Duarte, J., Veldhoen, M. et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 12, 255–263 (2011). https://doi.org/10.1038/ni.1993
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1993
This article is cited by
-
The choroid plexus acts as an immune cell reservoir and brain entry site in experimental autoimmune encephalomyelitis
Fluids and Barriers of the CNS (2023)
-
PD-L1 positive astrocytes attenuate inflammatory functions of PD-1 positive microglia in models of autoimmune neuroinflammation
Nature Communications (2023)
-
Cholinergic control of Th17 cell pathogenicity in experimental autoimmune encephalomyelitis
Cell Death & Differentiation (2023)
-
Growth hormone releasing hormone signaling promotes Th17 cell differentiation and autoimmune inflammation
Nature Communications (2023)
-
T cells: an emerging cast of roles in bipolar disorder
Translational Psychiatry (2023)