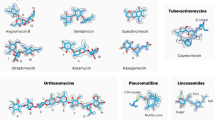
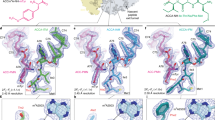
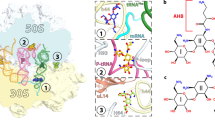
Abstract
Ribosomes, the site of protein synthesis, are a major target for natural and synthetic antibiotics. Detailed knowledge of antibiotic binding sites is central to understanding the mechanisms of drug action. Conversely, drugs are excellent tools for studying the ribosome function. To elucidate the structural basis of ribosome–antibiotic interactions, we determined the high-resolution X-ray structures of the 50S ribosomal subunit of the eubacterium Deinococcus radiodurans, complexed with the clinically relevant antibiotics chloramphenicol, clindamycin and the three macrolides erythromycin, clarithromycin and roxithromycin. We found that antibiotic binding sites are composed exclusively of segments of 23S ribosomal RNA at the peptidyl transferase cavity and do not involve any interaction of the drugs with ribosomal proteins. Here we report the details of antibiotic interactions with the components of their binding sites. Our results also show the importance of putative Mg+2 ions for the binding of some drugs. This structural analysis should facilitate rational drug design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Pioletti, M. et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20, 1829–1839 (2001).
Winberly, B. T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Schluenzen, F. et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 (2000).
Polacek, N., Gaynor, M., Yassin, A. & Mankin, A. S. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411, 498–501 (2001).
Thompson, J. et al. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl Acad. Sci. USA 98, 9002–9007 (2001).
Barta, A. et al. Mechanism of ribosomal peptide bond formation. Science 291, 203a (2001).
Spahn, C. M. T. & Prescott, C. D. Throwing a spanner in the works: antibiotics and the translation apparatus. J. Mol. Med. 74, 423–439 (1996).
Cundliffe, E. in The Ribosome: Structure, Function and Evolution (eds Hill, W. E. et al.) 479–490 (ASM, Washington DC, 1990).
Vester, B. & Douthwaite, S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45, 1–12 (2001).
Harms, J. et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell (in the press).
Vester, B. & Garrett, R. A. The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer center of Escherichia coli 23S ribosomal RNA. EMBO J. 7, 3577–3588 (1988).
Polacek, N. in RNA-binding Antibiotics (eds Schroeder, R. & Wallis, M. G.) 1–10 (Eurekah.Com, Georgetown, 2000).
Rodriguez-Fonseca, C., Amis, R. & Garrett, R. A. Fine structure of the peptidyl transferase centre on 23 S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol. 247, 224–235 (1995).
Moazed, D. & Noller, H. F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochemie 69, 879–884 (1987).
Mankin, A. S. & Garrett, R. A. Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon Halobacterium halobium. J. Bacteriol. 173, 3559–3563 (1991).
Izard, T. & Ellis, J. The crystal structures of chloramphenicol phosphotransferase reveal a novel inactivation mechanism. EMBO J. 19, 2690–2700 (2000).
Shaw, W. V. & Leslie, A. G. W. Chloramphenicol acetyl-transferase. Annu. Rev. Biophys. Biophys. Chem. 20, 363–386 (1991).
Kalliaraftopoulos, S., Kalpaxis, D. L. & Coutsogeorgopoulos, C. New aspects of the kinetics of inhibition by lincomycin of peptide-bond formation. Mol. Pharmacol. 46, 1009–1014 (1994).
Douthwaite, S. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal-RNA. Nucleic Acids Res. 20, 4717–4720 (1992).
Vester, B., Hansen, L. H. & Douthwaite, S. The conformation of 23S ribosomal-RNA nucleotide A2058 determines its recognition by the Erme methyltransferase. RNA 1, 501–509 (1995).
Ross, J. I. et al. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob. Agents Chemother. 41, 1162–1165 (1997).
Steiner, G., Kuechler, E. & Barta, A. Photo-affinity labeling at the peptidyl transferase center reveals 2 different positions for the A-site and P-sites in domain-V of 23S ribosomal-RNA. EMBO J. 7, 3949–3955 (1988).
Vazquez, D. Inhibitors of Protein Synthesis (Springer, Berlin, 1975).
Yonath, A., Leonard, K. R. & Wittmann, H. G. A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science 236, 813–816 (1987).
Milligan, R. A. & Unwin, P. N. Location of exit channel for nascent protein in 80S ribosome. Nature 319, 693–695 (1986).
Steinmetz, W. E., Bersch, R., Towson, J. & Pesiri, D. The conformation of 6-methoxyerythromycin-A in water determined by proton NMR-spectroscopy. J. Med. Chem. 35, 4842–4845 (1992).
Gasc, J. C., Dambrieres, S. G., Lutz, A. & Chantot, J. F. New etheroxime derivatives of erythromycin A: a structure–activity relationship study. J. Antibiot. 44, 313–330 (1991).
Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585 (1995).
Sigmund, C. D., Ettayebi, M. & Morgan, E. A. Antibiotic-resistance mutations in 16S and 23S ribosomal-RNA genes of Escherichia coli. Nucleic Acids Res. 12, 4653–4663 (1984).
Bottger, E. C., Springer, B., Prammananan, T., Kidan, Y. & Sander, P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2, 318–323 (2001).
Goldman, R. C., Fesik, S. W. & Doran, C. C. Role of protonated and neutral forms of macrolides in binding to ribosomes from gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 34, 426–431 (1990).
Mao, J. C.-H. & Puttermann, M. The intermolecular complex of erythromycin and ribosome. J. Mol. Biol. 44, 347–361 (1969).
Poulsen, S. M., Kofoed, C. & Vester, B. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304, 471–481 (2000).
Hansen, L. H., Mauvais, P. & Douthwaite, S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31, 623–631 (1999).
Xiong, L. Q., Shah, S., Mauvais, P. & Mankin, A. S. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31, 633–639 (1999).
Wittmann, H. G. et al. Biochemical and genetic studies of two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 127, 175–189 (1973).
Gregory, S. T. & Dahlberg, A. E. Erythromycin resistance mutation sin ribosomal proteins L22 and L4 perturb the high order structure of 23S ribosomal RNA. J. Mol. Biol. 289, 827–834 (1999).
Saarma, U., Spahn, C. M. T., Nierhaus, K. H. & Remme, J. Mutational analysis of the donor substrate binding site of the ribosomal peptidyltransferase center. RNA 4, 189–194 (1998).
Moazed, D. & Noller, H. F. Sites of interaction of the CCA-end of peptidyl-transfer RNA with 23S ribosomal-RNA. Proc. Natl Acad. Sci. USA 88, 3725–3728 (1991).
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Vince, R., Almquist, R. G., Ritter, C. L. & Daluge, S. Chloramphenicol binding site with analogues of chloramphenicol and puromycin. Antimicrob. Agents Chemother. 8, 439–443 (1975).
Tenson, T., DeBlasio, A. & Mankin, A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl Acad. Sci. USA 93, 5641–5646 (1996).
Noll, M., Hapke, B., Schreier, M. H. & Noll, H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J. Mol. Biol. 75, 281–294 (1973).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Murshudov, G. N., Lebedev, A., Vagin, A. A., Wilson, K. S. & Dodson, E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999).
Carson, M. Ribbons. Methods Enzymol. 277, 493–505 (1997).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Acknowledgements
We thank A. Hofmann for advice; A. Mankin and F. Triana for critical discussions; and T. Auerbach, H. Bartels, W. S. Bennett, H. Burmeister, C. Glotz, M. Glühmann, H. A. S. Hansen, M. Kessler, M. Laschever, S. Meier, J. Muessig, M. Peretz, M. Pioletti, B. Schmidt, A. Sitka, C. Stamer and A. Vieweger for contributing to different stages of these studies. These studies could not have been performed without the cooperation of the staff of the synchrotron radiation facilities at EMBL and Max Planck Gesellschaft at DESY, ID14/2&4 at EMBL/ESRF and ID19/APS/ANL. Support was provided by the Max-Planck Society, the US National Institute of Health, the German Ministry for Science and Technology and the Kimmelman Center for Macromolecular Assembly at the Weizmann Institute. A.Y. holds the Martin S. Kimmel Professorial Chair.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schlünzen, F., Zarivach, R., Harms, J. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001). https://doi.org/10.1038/35101544
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35101544
This article is cited by
-
The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services
Nature Communications (2023)
-
Ribosome biogenesis in disease: new players and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Can intracellular Staphylococcus aureus in osteomyelitis be treated using current antibiotics? A systematic review and narrative synthesis
Bone Research (2022)
-
Structural and functional insights into esterase-mediated macrolide resistance
Nature Communications (2021)
-
Context-specific action of macrolide antibiotics on the eukaryotic ribosome
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.