Abstract
Introduction
Surveillance systems for acute respiratory infections (ARI) in children currently are often limited in terms of the panel of pathogens and the age range investigated or are only syndromic and at times only active in the winter season.
Methods
Within PID-ARI.net, a research network for ARI in children in Germany, an active, year-round surveillance system was formed in three regions from north to south for population-based analysis. Children from birth to 16 years of age were included and up to 19 noncolonizing airway pathogens were tested for with multiplex RT-PCR.
Results
In the 10-year period from July 1996 to June 2006, a total of 18,899 samples were tested. The positive rate increased with the size of the test panel to up to 72.9%. Picornaviruses (35–39%), paramyxoviruses (23–28%) and orthomyxoviruses (5.8–12.5%) comprised the highest fraction. Reoviruses and Legionella pneumophila were not found at all and Chlamydia pneumoniae and Bordetella parapertussis only rarely. Respiratory syncytial virus and parainfluenza virus (PIV) type 3 were anticyclical in rhythmicity with metapneumovirus and PIV1 and PIV2. The age medians per pathogen depended predominantly upon the attack rate and interepidemic intervals.
Conclusion
Active surveillance systems for ARI are superior to passive systems. They should be pathogen-specific and comprehensive for viruses and bacteria and age ranges. They should be population-based and multilevel to avoid bias. The impact of atypical bacteria in children was highly overestimated in earlier studies.
Similar content being viewed by others
Introduction
Pathogen-specific surveillance, as opposed to syndromic surveillance, such as monitoring for “influenza-like illness (ILI),” is the key to elucidating the epidemiology of ARI in children systematically and in detail [34]. Since laboratory confirmation of an episode of ARI is not systematically undertaken in most health care systems, active surveillance systems have to be created to ascertain a sufficient number of cases to yield high quality data. So far, most systems have only focused on a very limited number of pathogens or a limited age range of children and have been used under circumstances in which the denominator of the investigation remained unclear or was not sufficiently characterized. Since budgets are limited, active surveillance systems have to be limited in size. Different segments of the pediatric population such as hospitalized children, outpatients or population cohorts can be investigated. Until recently, several very different laboratory techniques and cell lines for different types of bacteria and viruses, respectively, had to be used to cover most of the known ARI pathogens [13, 29].
Methods
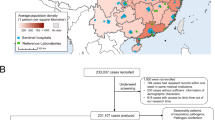
The described surveillance system started in July 1996 in Kiel and later on was the key element of the research network PID-ARI.net (http://www.pid-ari.net) [34]. It was active in the form and size described here until the end of June 2006, a 10-year period. Since the calendar year is an unsuitable time frame for ARI, an epidemiological year (ey) was defined as July to the end of June the following year. An even ey starts with July of an even calendar year (e.g., 2004/2005) and an uneven ey with July of an uneven calendar year (e.g., 2005/2006). In three defined regions, all hospitals treating children were included to form an epidemiological recruitment system (ERS) for population-based analysis: north–University Children’s Hospital Kiel, Municipal Hospitals Kiel and Flensburg; mid–University Children’s Hospital Mainz; south–University Children’s Hospital Freiburg and Municipal Hospital Freiburg. The University Children’s Hospital Mainz serves as a third-degree referral center as well as a municipal hospital. Pediatric offices in each region were included. They chose one day of the week in which they had to enroll all patients fulfilling the inclusion criteria. Included were all LRI and complicated ARI (Fig. 1). The same categories were applied as in earlier reports [35].
Naso-pharyngeal aspirates (NPA) were obtained from the recruited children according to a standard operating protocol and sent to the central network laboratory in Kiel on Mondays, Tuesdays and Wednesdays by surface mail or stored at 2–8°C until the following Monday. In Kiel, the samples were tested with a 9-valent multiplex RT-PCR-ELISA until October 2002, then until April 2003 with a 16-valent and from then onwards with a 19-valent technique [6, 25, 34]. Most of the known upper-airway noncolonizing pathogens were included in the test kit (Table 1). Primers referring to the following regions or proteins were chosen: influenza A (IVA)—nonstructural protein, primers detecting both H3N2 and H1N1; influenza B (IVB)—nonstructural protein; respiratory syncytial virus (RSV)—fusion protein F1 subunit, detecting groups A and B; human metapneumovirus (MPV)—nucleoprotein; parainfluenza type 1 (PIV1)—hemagglutinin neuraminidase; parainfluenza type 2 (PIV2)—nucleocapsid protein; parainfluenza type 3 (PIV3)—hemagglutinin neuraminidase; parainfluenza type 4 (PIV4)—phosphoprotein detecting groups A and B; coronavirus types OC43 (CV OC43)—open reading frame 1b of the polymerase gene and matrix protein, and 229E (CV 229E)–polymerase gene; picornaviruses—5′ noncoding region; rhinovirus (RV) and enterovirus (EV)—5′ noncoding region; reoviruses (ReoV)—minor core protein lambda 3 (L1 gene) detecting groups 1, 2, and 3; adenovirus (AV)—hexon protein; Mycoplasma pneumoniae (Mpn)—16s-rRNA; Chlamydia pneumoniae (Cpn)—16s-rRNA; Bordetella pertussis (Bp)—IS481; Bordetella parapertussis (Bpp)—porin gene; and Legionella pneumophila species (Lpn)—region of 23srRNA and 5srRNA.
Conventional bacteria, such as pneumococci or staphylococci, which do colonize the upper airways were not included in the panel because samples from the upper airways, such as an NPA, cannot prove the etiology of an LRI.
MS Access served as the software for data bank management. SPSS version 10.0 was used for biostatistics. Since the age distributions were mostly considerably skewed, the Wilcoxon rank-sum test and Mann-Whitney U-test for two independent samples were used as nonparametric tests to investigate the difference between the median ages.
Results
From July 1996 to June 2006, a total of 18,899 samples were tested (Table 1), of which 16,423 (86.9%) were from inpatients and 2,476 (13.1%) from outpatients. The median age (mean) was 22 months (40 months) for all children, 21 months (39 months) for inpatients and 41 months (50 months) for outpatients. The median difference of 20 months between in- and outpatients was highly significant (P < 0.001).
Ascertainment of NPA was related to age and increased with younger age from 64.0% in children over 5 years to 78.4% in infants. The diagnoses were 25.0% complicated upper ARI, 3.2% laryngo-tracheo-bronchitis, 9.7% bronchitis (nonwheezing), 24.8% wheezing bronchitis including bronchiolitis, 24.5% bronchopneumonia, 13.2% pneumonia and 11.0% non-ARI. The latter did not fulfill the inclusion criteria, but the ERS doctors used the procedure in the hope of finding patients with other disease conditions.
Positive rate
The positive rate increased with the number of pathogens in the test kit (Table 1). If the non-ARI patients (11%) with a lower yield are subtracted, the positive rate for the 19-valent technique reached 72.9%. During the 9-valent phase, the positive rate was heavily influenced by the severity of the RSV season. This effect was neutralized after including MPV and RV in the m-RT-PCR. The positive rates per region were north 62.8%, mid 63.2%, south 67.7% (chi-squared 24.5, df = 2, P < 0.001, test for trend P = 0.045).
ReoV and Lpn were not found at all. Multiple infections were present in 12.5% (only data for extended panels from October 2002 to July 2006). The fractions of co-infections per pathogen were IVA 17.8%, IVB 19.9%, RSV 25.3%, MPV 32.4%, PIV1 37.0%, PIV2 36.6%, PIV3 28.7%, PIV4 43.4%, CV OC43 and 229E 59.3%, RV 26.4%, EV 36.2%, AV 53.1%, Mpn 40.5%, Cpn 46.2%, Bp 60.8%, and Bpp 81.8%. The orthomyxoviruses were least involved in co-infections. The co-infection rate within the paramyxoviruses was higher for PIV1 (P = 0.049) and PIV4 (P = 0.005) and not statistically significantly different for the others.
Single pathogens
RSV occurred annually, and most years went into a so-called epidemiological eclipse in the summer months (Fig. 2a). In the summer of 2002, an outbreak in the coastal area of Schleswig-Holstein (north) took place. In 2004, sporadic cases were seen during the summer. In uneven ey, the RSV season started late at the end of December to January and was less severe than in even ey when the season started early at the end of September to October with a high incidence. In severe RSV seasons, the attributable fraction of RSV in infants was as high as 46.7%.
a Monthly incidence of pathogens RSV, PIV1, PIV3, IVA, IVB, EV, and AV from July 1996 to June 2006 and PIV2, PIV4, MPV, and RV from October 2002 and CV from April 2003 onwards only. b Monthly incidence of pathogens Mpn and Cpn from July 1996 to June 2006 and Bp and Bpp from October 2002 onwards. Note that the scale on the vertical axis differs between a and b. See Table 1 for abbreviations
PIV3 is endemic with epidemics mainly in the spring months. The incidence of PIV3 seems to parallel the RSV years, but contributes only about one-fifth of the burden of RSV (Table 1).
MPV is the second most burdensome disease in terms of incidence within the paramyxoviruses. It appears anticyclical with RSV, and early seasons are also severe and late seasons weak in incidence. The cycles and rhythm of MPV were confirmed by retrospective testing of stored material through 1996 (data not shown), since the virus was only discovered in 2001 [31].
PIV1 and PIV2 occur in epidemic proportions biennially in uneven ey with an early start in late August to September, with PIV1 always higher in incidence than PIV2. Within the group of paramyxoviruses, PIV1 and 2, along with MPV, fill in for weak RSV seasons. For many weeks, MPV, PIV1 and PIV2 can be the dominating pathogens especially in uneven ey. The incidence of RSV then only increases once these pathogens decline. As mentioned above, this happens every other year. All paramyxoviruses together contributed about 25% (22.6–27.5%) to the total disease episodes in children up to 16 years of age (Table 1). PIV4 seems to be the least important paramyxovirus. After three active seasons since 2002, there was no activity of this virus in epidemic proportions in 2005/2006. Therefore no clear rhythm can be noted yet.
The impact of IVA was higher than IVB in all seasons except the most recent one. The attributable fraction of influenza viruses varied from 0.8% in the weak season 2005/2006 to 10.4% in the most severe season 2002/2003. IVA occurred annually with a clear epidemiological eclipse in the summer months. The IVA seasons started between calendar weeks 47 and 5. IVB does not occur in epidemic proportions every year. One- to two-year gaps such as in ey 2003/2004, and 1999/2000–2000/2001 were observed. In 2001/2002 and 2002/2003, IVB occurred in 2 consecutive years.
Within the picornaviruses, RV showed a clear preponderance over EV. Both together contributed up to 38.4%. Both viruses are endemic all year, and the incidence increases in late summer. Other viruses such as RSV may have a higher incidence only during a few months of the year.
The seasonality of CV OC43 and CV 229E together follows an annual epidemic pattern. In a given year, either CV OC43 or CV 229E was predominant (data not shown). Coronaviruses are the viruses within the panel that are most often involved in co-infections (59.3%).
AV, the only DNA virus in the panel, was endemic all year throughout the complete observation period with a higher incidence in the winter months. AV is also highly involved in co-infections (53.1%).
The contribution of Mpn appeared marginal until the last season, 2005/2006, when a widespread epidemic was observed in Germany (Fig. 2b). After 1998/1999, it took 7 years for another widespread Mpn outbreak to occur. Only in 33 cases was Cpn diagnosed, and a second pathogen was detected in almost half of the cases (46.2%).
The highest incidence of Bp observed so far occurred in 2004/2005 with a 1.0% fraction. There were, however, only 35 positive samples, and half were involved in co-infections (48.1%). Bpp was even lower in incidence. Only 22 cases were observed in 4 years, and most cases were involved in co-infections (81.6%).
Epidemiological pattern
The epidemiological pattern was “epidemic yearly” for RSV, MPV, IVA, and CV; “epidemic biennially” for PIV1 and PIV2; “epidemic irregularly” for IVB and PIV4; “endemic epidemic” for PIV3, AV, RV, EV, Mpn, and Bp; and “sporadic” for Bpp and Cpn (Fig. 2a,b).
Median age
Figure 3 shows the median age of the children by pathogen, stratified by inpatient versus outpatient status. Since the age distribution was considerably skewed, the Wilcoxon rank-sum test was used as nonparametric test for the difference in the medians. RSV is particularly successful in attacking children considerably earlier than the other paramyxoviruses if one considers the inpatients. The median age of PIV3-positive outpatients (n = 74) was the lowest, and it was only slightly higher than in inpatients (see legend to Fig. 3). No statistical difference in median age between in- and outpatients existed for PIV4, Bpp, Mpn and Cpn. The hospitalized Bp-positive patients were very young.
Median ages of patients diagnosed with different pathogens stratified by inpatients and outpatients. Significance of the differences of the median: P(RSV)<0.001, P(PIV1)=0.002, P(PIV2)=0.524, P(PIV3)=0.049, P(PIV4)=0.062, P(MPV)<0.001, P(IVA)<0.001, P(IVB)=0.272, P(EV)<0.001, P(RV)<0.001, P(CV)=0.001, P(AV)<0.001, P(Bp)=0.001, P(Bpp)=0.695, P(Mpn)=0.331, P(Cpn)=0.598 (For abbreviations, see Table 1)
Discussion
The system
The surveillance system at the given size with three geographical regions from northern to southern Germany was intended to be a basis for a research network on ARI in children [34]. By covering the children of 2% of the total population living in Germany and the three areas, the system is of the sentinel type. The size of the system was limited by the financial resources available. This had an effect on the detection of pathogens that cause more local outbreaks, such as Mpn, Cpn and Lpn, that therefore could be missed by the system; however, the 10-year period should have provided the system with sufficient power to detect these pathogens if they occurred in sufficient numbers across a considerable geographic spread. For pathogens with widespread activity, such as most viruses in their main seasons, the limited geographical size should have had no influence.
Given the limited resources, the system focused on the tip of the ARI iceberg, i.e. hospitalized children. Hospitalization is an acceptable surrogate for severe ARI. Inpatients are a selected subpopulation and are not necessarily representative of children in general for this reason. To broaden the picture, outpatients were included. Ascertainment of NPA in our system was clearly related to age and increased with younger age. This biased the results toward pathogens related to a younger age, as the sample was dominated by younger age as 86.9% of the patients were recruited in hospital. This can be seen by the far right skewing of the age distributions (see medians and means given). Young age is a known risk factor for hospitalization [5]. Therefore the data were stratified by hospitalization status.
Diagnostics
A bias towards more resistant/viable pathogens because of transportation of the samples is unlikely since the positive rate measured by m-RT-PCR did not decrease with distance to the diagnostic laboratory, but was inverse as the laboratory was located in the north region. PCR, especially multiplex RT-PCR is a recent, commonly used technique for this type of investigation [9, 36] that does not need viable pathogens but a sufficiently well-preserved genome. This saves considerable funds since no special cooling containers have to be used, and simplicity is a very important aspect to sustaining such a system over a long period of time.
Validation of m-RT-PCRs is a major issue and requires a considerable amount of effort, particularly if one has high standards in terms of sample size and gold-standard testing [25]. Before October 2002, the sensitivity of the m-RT-PCR for PIV3 was poor (Fig. 2a) as the primers published by Karron et al. [15] detected only about 60% of PIV3-positive findings. After changing the primers from the F-gene to the hemagglutinin-neuraminidase gene, sensitivity reached the desired level [25]. Delineation between EV and RV is difficult since both viruses are closely related, and therefore some misjudgment could have occurred. This, however, should not have compromised the interpretation considerably, since the two viruses are closely related. Data from Finland demonstrated that EV is genetically more closely related to group B RV than group A is to group B RV, and the new name enterorhinovirus was proposed to indicate that the distinction is artificial [17].
Co-infections became an issue only after molecular diagnostic techniques, especially PCR technology, facilitated the co-identification of pathogens [12]. The 12.5% of our data result from the size of the panel investigated, especially regarding viruses commonly found in co-infections, age and recruitment type (i.e., in- versus outpatients). A preponderance of RSV together with MPV as found by the group in Liverpool in severe infections could not be confirmed by our data [28]; however, the detailed clinical data are still under evaluation. Length of time of persistence of a given pathogen in the host and a less severe clinical course (outpatients) increased the probability of acquiring a second pathogen or a co-infection before referral for medical treatment. So far, the chance of a co-infection appears more stochastical than representative of a specific relationship between two pathogens. Sequential infections of viruses and bacteria are well known [20, 24], but conventional bacteria were not included in our test panel for the reasons mentioned above.
Single pathogens, rhythmicity, severity
The dual rhythm of RSV is well known [3, 32], and more recently it has also been accepted in the United States [22]. This rhythm was also well modeled by Weber et al. [33]. Climatic factors or alternating subtypes are an unlikely explanation for alternating early and late seasons. An interesting explanation would be the interference of other viruses such as parainfluenza types 1 and 2 as mentioned by Glezen [4]. Our discovery of MPV as the major competitor of RSV further substantiates this hypothesis. For viruses with a distinct rhythmicity, clinically important predictions can currently be made at least for RSV, e.g., regarding curtailing passive immunization with palivizumab [34]. During the last 7 years, we were able to predict the onset of RSV correctly. That RSV in some years does not go into an epidemiological eclipse was also observed in coastal areas of the United States [7]. Whether sporadic positive samples for epidemic viruses can be found in the population depends on the sample size investigated and the age range of the surveillance system. Most recently, adult chronic obstructive pulmonary disease (COPD) patients have been considered as a reservoir for RSV [37]. In addition, premature infants excrete the virus over many months.
The epidemic onset of IVA in children varies over 9 weeks and does not show a clear pattern. Also the strength of the season cannot be predicted since it depends on how far the current strains have drifted from previous ones and on the severity of the previous seasons [1]. The Fujian variant in 2003/2004, for example, which caused much upset in the United Kingdom and Ireland (see http://www.eurosurveillance.org, 2003), did not have a major impact in Germany, since the preceding year had been a severe season with closely related ancestral strains in Germany. The 2005/2006 epidemic was particularly weak, most likely for similar reasons and the highest vaccine use in the population so far. Szucs reported 33% vaccine coverage rate for the total population of Germany in the season 2005/2006, a coverage expected to break an influenza epidemic (T.D. Szucs, personal communication).
IVB occurs irregularly as observed in the German national (AGI) and European surveillance system (EISS). The reason that IVB occurred in epidemic proportions in 2 consecutive years was that the Yamagata lineage was followed by a Victoria lineage, and because this strain had not circulated for many years, no immunity existed in children. Both lineages have been circulating since then.
The high impact of enterorhinovirus described in this study was confirmed by others [16, 18]. RV so far has been proven to cause LRI in addition to upper ARI. Papadopoulos et al. could clearly show that RV can replicate very well in lower respiratory tract epithelium [23]. More and more groups describe the considerable impact of RV in hospitalized ARI cases and in ARI in children in general. In the meantime, RV is regarded as more important for lung health and reactive airway disease in children than paramyxoviruses such as RSV [11]. EV can persist for long periods in the host, and therefore patients are prone to co-infections with other pathogens, which makes estimation of attributable fractions more complicated or impossible. In this respect, our data show that EV is somehow distinct from RV.
Coronaviruses are also not thought to cause LRI in a similar way to RV. Their contribution has most likely been underestimated. A real boost for CV research came with SARS in 2003 [14]. Only in 2004 were two new types of CV detected, CV-NL63 and CV-HKU1, and they were not included in our test panel.
AV can persist in the tonsils for many months and therefore is very often involved in co-infections, making a statement on its role in the individual patient difficult [10]. AV and pathogens frequently involved in co-infections are more likely to be the responsible pathogen, if they are detected as the only pathogen.
Mpn occurs more frequently in local outbreaks than in widespread epidemics. The 1998/1999 epidemic in Denmark was also paralleled in our northern region (Andersen PH, EPI-News 46/1998, http://www.ssi.dk). Earlier data from Denmark showed a changing epidemiological pattern [19]. In the last ey of the observation period, the highest Mpn incidence so far was registered in the northern and southern regions. In Denmark, the most recent widespread epidemic was reported in the ey 2004/2005, i.e., 1 year earlier (Andersen PH, EPI-News 12/2005, http://www.ssi.dk).
The impact of Cpn based on sero-epidemiology has so far been highly over-estimated [27]. The same level of incidence as reported by us was demonstrated by CAPNETZ, another German research network on community-acquired pneumonia in adults (data presented at the meeting of the Deutschen Gesellschaft für Mikrobiologie und Hygiene 2005) and an extensive study based on PCR analysis from Greece revealed no cases [30]. Incorrectly assigned data regarding a high burden of disease for Mpn and Cpn implied a widespread macrolide use in pediatrics in Germany. This had severe consequences for medical practice, since macrolide-resistance in pneumococci is increasing to worrying proportions of over 35% of children under 5 years in Germany [26]. Arguments that the m-RT-PCR is not sensitive enough or that NPA is an unsuitable sample for detecting Cpn as an intracellular pathogen within epithelial cells, which are present in lower numbers in NPA compared to swab samples, can be refuted [35]. In the validation process, the 9v PCR was comparable to other PCRs and standard techniques [25].
After three of the major trials of acellular pertussis vaccines were carried out in Germany in the late 1980s, the acellular vaccines were licensed in 1991 and since then have been used in the routine vaccination program. This increased the vaccine uptake rate to over 90%, and the incidence of pertussis declined leaving only young infants in the pre-vaccination age unprotected. This is the most likely explanation for the wide divide between in- and outpatients in median age.
From data of the pre-vaccination era, it is known that Bpp has a lower impact than Bp; however, the currently used acellular vaccines also have a suppressive effect on Bpp, at least as demonstrated by a vaccine efficacy of 31% for the five-component Lederle vaccine, following a Bpp community outbreak in the study area where the vaccine was tested [8]. This could explain a similar effect on the median age between in- and outpatients as seen for Bp. The difference in the median age for Bpp, however, was not statistically significant due to the small number of 22 patients.
Lpn is not a pediatric pathogen according to our data. Up to now, only a few case reports, mainly of nosocomial infections in children, have been published. This statement is confirmed by a recent outbreak in France in 2003/2004, when Lpn was spread over vast areas of northern France by a contaminated cooling tower of an industrial power station [21]. In this outbreak, 86 cases were confirmed with the youngest one being 32 years of age and an attack rate that increased with age.
Since rotavirus, another reovirus, is a major cause of diarrhea in children and only a few data on respiratory reoviruses exist so far, primers for groups 1, 2, and 3 reoviruses were included in the panel. As not a single sample tested positive in the three seasons in which the 19-valent PCR was used, it is unlikely that reoviruses have a major impact in pediatric ARI. The PCR, however, could only be tested on one sample per reovirus group for reasons of availability.
Median age
The basic reproductive coefficient (Ro), attack rate, interepidemic interval, antigenic variability of a given pathogen and the immunity transferred by the mother and maintained in the offspring after infection are important variables influencing the median age of infection and disease. Ro, which can be estimated as the quotient of life expectancy divided by average age of first infection, was the highest in RSV and PIV3. The lower the attack rate per year and the wider the interepidemic interval, the higher the median age. The “subtype phenomenon” described earlier [35], refers to IVA and IVB, when children become older with a nonprotective immunity because the involved strains have drifted far apart and therefore a potential cross-protection does not occur [1]. This would imply an effect similar to being infected initially with a stable virus at a later age. It is most likely true of all ARI pathogens that first infections are more likely to be severe ARI or LRI than infections later in life when the host has already experienced previous infections with the pathogen and upper ARI become more likely. The fact that the median age of inpatients for Mpn and Cpn was similar to outpatients alludes also to the influence of the attack rate and interepidemic interval and possibly to the immune-enhancement conferred by earlier infections, a well-described phenomenon in Mpn infections [2].
Limitations
Analysis costs are a limitation, but they are low in contrast to conventional tests for this number of pathogens. Since conventional bacteria were not included in the test panel for lack of availability of practical diagnostic options in LRI in children, this diagnostic dilemma limits the final delineation of attributable fractions and therefore also causality.
Conclusions
Population-based surveillance with a known denominator and source population is a prerequisite for valid epidemiological research in ARI, otherwise investigations are prone to bias. Surveillance data generated by a well-characterized recruitment system and a comprehensive test panel are less vulnerable to bias. Comprehensive and continuously updated panels of pathogens tested by multiplex RT-PCR are desirable. Picornaviruses, paramyxoviruses and orthomyxoviruses are of highest impact in ARI in children. Orthomyxoviruses are less frequently involved in co-infections than other pathogens within the panel. Bpp, Cpn, ReoV and Lpn are not important pathogens of acute ARI in childhood. For viruses with regular epidemic patterns, predictions can be made.
Abbreviations
- ARI:
-
acute respiratory infection
- ERS:
-
epidemiological recruitment system
- ey:
-
epidemiological year
- LRI:
-
lower respiratory infections
- m-RT-PCR:
-
multiplex reverse transcription polymerase chain reaction
- NPA:
-
nasopharyngeal aspirate
References
Boni MF, Gog JR, Andreasen V, Christiansen FB (2004) Influenza drift and epidemic size: the race between generating and escaping immunity. Theor Popul Biol 65:179–191
Broughton RA (1986) Infection due to Mycoplasma pneumoniae in childhood. Pediatr Infect Dis J 5:71–85
Florman AL, McLean LC (1988) The effect of altitude and weather on the occurrence of outbreaks of respiratory syncytial virus infections. J Infect Dis 158:1401–1402
Glezen WP, Denny FW (1973) Epidemiology of acute lower respiratory disease in children. N Engl J Med 288:498–505
Graham NMH (1994) Respiratory infections. In: Pless IB (ed) The epidemiology of childhood disorders. Oxford University Press, New York pp 173–210
Gröndahl B, Puppe W, Hoppe A, Kühne I, Weigl JAI, Schmitt HJ (1999) Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol 37:1–7
Halstead D, Jenkins SG (1998) Continuous non-seasonal epidemic of respiratory syncytial virus infection in the Southeast United States. South Med J 91:433–436
Heininger UH, Stehr K, Christenson P, Cherry JD (2004) Evidence of efficacy of the Lederle/Takeda acellular pertussis component diphtheria and tetanus toxoids and pertussis vaccine but not the Lederle whole-cell component diphtheria and tetanus toxoids and pertussis vaccine against Bordetella parapertussis infection. Clin Infect Dis 28:602–604
Hendrickson KJ, Hoover S, Hehl KS, Hua W (2004) National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis J 23:S11–S18
Horwitz MS (1996) Adenoviruses. In: Fields BN, Knipe DM, Howley PM, Channock RM, Melnick JL, Monath TP, Roizman B, Straus SE (eds) Fields virology, 3rd ed. vol 2. Lippincott-Raven, Philadelphia, pp 2149–2171
Jartti T, Lehtinen P, Vuorinen T, Osterback R, van den Hoogen B, Osterahus AD, Ruuskanen O (2004) Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis 10:1095–1101
Jennings LC, Anderson TP, Werno AM, Beynon KA, Murdoch DR (2004) Viral etiology of acute respiratory tract infections in children presenting to hospital. Role of polymerase chain reaction and demonstration of multiple infections. Pediatr Infect Dis J 23:1003–1007
Juven T, Mertsola J, Waris M, Leinonen M, Meurman O, Roivainen M, Eskola J, Saikku P, Ruuskanen O (2000) Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J 19:293–298
Kahn JS, McIntosh K (2005) History and recent advances in coronavirus discovery. Pediatr Infect Dis J 24:S223–S227
Karron RA, O’Brien KL, Froehlich JL, Brown VA (1993) Molecular epidemiology of a parainfluenza type 3 virus outbreak on a pediatric ward. J Infect Dis 167:1441–1445
Kellner G, Popow-Kraupp T, Kundi M, Binder C, Wallner H, Kunz C (1988) Contribution of rhinoviruses to respiratory viral infections in childhood: a prospective study in a mainly hospitalized infant population. J Med Virol 25:455–469
Laine P, Savolainen C, Blomqvist S, Hovi T (2005) Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol 86:697–706
Legg JP, Warner JA, Johnston S, Warner JO (2005) Frequency of detection of picornaviruses and seven other respiratory pathogens in infants. Pediatr Infect Dis J 24:611–616
Lind K, Bentzon MW (1988) Changes in the epidemiological pattern of Mycoplasma pneumoniae infections in Denmark–a 30 years survey. Epidemiol Infect 101:377–386
Madhi SA, Klugmann KP, the Vaccine Trialist Group (2004) A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med 8:811–813
Minh NNT, Ilef D, Campese C, Che D, Ganiayre F, Haeghebaert S, Guitard C, Panie G, Jarraud S, Marcel F, Desenclos JC (2004) A prolonged outbreak of Legionnaires’ disease associated with an industrial cooling tower–how far can airborne transmission spread? Abstracts of the 9th EPIET Scientific Seminar, Mahon, Menorca, October 14–16, p 57
Mullins JA, Lamonte AC, Bresee JS, Anderson LJ (2003) Substantial variability in community respiratory syncytial virus season timing. Pediatr Infect Dis J 22:857–863
Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, Fraenkel DJ, Meyer J, Lackie PM, Sanderson G, Holgate ST, Johnston SL (2000) Rhinoviruses infect the lower airways. J Infect Dis 181:1875–1884
Peltola VT, Murti KG, McCullers JA (2005) Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 192:249–257
Puppe W, Weigl JAI, Aron G, Gröndahl B, Schmitt HJ, Niesters HGM, Groen J (2004) Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J Clin Virology 30:165–174
Reinert RR, Luetticken R, Brvskier A, Al-Lahham A (2003) Macrolide-resistant Streptococcus pneumoniae and Streptococcus pyogenes in the pediatric population in Germany during 2000–2001. Antimicrobial Agents Chemoth 47:489–493
Schmidt SM, Müller CE, Mahner B, Wiersbitzky SKW (2002) Prevalence, rate of persistence and respiratory tract symptoms of Chlamydia pneumoniae infections in 1211 kindergarten and school age children. Pediatr Infect Dis J 21:758–762
Semple MG, Cowell A, Dove W, Greensill J, McNamara PS, Halfhide C, Shears P, Smyth RL, Hart CA (2005) Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 191:382–386
Trollfors B, Claesson BA (1997) Childhood pneumonia: possibilities for aetiological diagnosis. Baillière’s Clin Paediatr 5:71–83
Tsolia MN, Psarras S, Bossios A, Audi H, Palanius M, Gourgiotis D, Kallergi K, Kafetzis DA, Constantopoulos A, Papadopoulos NG (2004) Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis 39:681–686
Van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD (2001) A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724
Waris M (1991) Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J Infect Dis 163:464–469
Weber A, Weber M, Milligan P (2001) Modeling epidemics caused by respiratory syncytial virus (RSV). Math Biosci 172:95–113
Weigl JAI, Puppe W, Meyer CU, Berner R, Forster J, Schmitt HJ, Zepp F, PID-ARI.net (2005) RSV-prevention in children guided by a web-based early warning system. Klin Pädiatrie 217:47–52
Weigl JAI, Puppe W, Gröndahl B, Schmitt HJ (2000) Epidemiological investigation of nine respiratory pathogens in hospitalized children in Germany using multiplex reverse transcriptase polymerase chain reaction. Eur J Clin Microbiol Infect Dis 9:336–343
Weinberg GA, Erdman D, Edwards KM, Hall CB, Walker FJ, Griffin MR, Schwartz B, the New Vaccine Surveillance Network Study Group (2004) Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis 189:706–710
Wilkinson TMA, Donaldson GC, Johnston SL, Openshaw PJM, Wedzicha JA (2006) Respiratory syncytial virus, airway inflammation, and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 73:871–876
Acknowledgements
We appreciate the help of our co-workers S. Rockahr, B. Reckewitz and G. Delfs. We thank Dr. von Klinggräff and PD Dr. Claaß, former and current head, respectively, of the Pediatric Department, Municipal Hospital, Kiel; PD Dr. Oldigs, head of the Department of Pediatrics, Municipal Hospital, Flensburg; and office pediatricians Mrs. Dr. and Mr. Schäfer, Izehoe; Dr. Hake and Dr. Morf, Flensburg; Dr. Kelber, Lüneburg; Dr. Neufang, Dr. Zaum, Freiburg; Dr. Eppinger, Bad Krozingen; and Dr. Vogel, Dr. Huber, Dr. Humber and Dr. Homann for their collaboration. We thank Dr. Adrian Sewell, University Children’s Hospital Frankfurt/Main, for linguistic assistance. This work was supported by the grant (FKZ 01KI0213) for PID-ARI.net, the Pediatric Infectious Diseases Network on Acute Respiratory Tract Infections, awarded by the Bundesministerium für Forschung (BMBF) via the Deutsches Zentrum für Luft- und Raumfahrt e.V.
Author information
Authors and Affiliations
Consortia
Corresponding author
Rights and permissions
About this article
Cite this article
Weigl, J.A.I., Puppe, W., Meyer, C.U. et al. Ten years’ experience with year-round active surveillance of up to 19 respiratory pathogens in children. Eur J Pediatr 166, 957–966 (2007). https://doi.org/10.1007/s00431-007-0496-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-007-0496-x